Highlights
- Soil pH and Cd speciation affect soil bacterial taxonomy and functions.
- Bacterial functional genes are sensitive to lime, biochar and straw amendments.
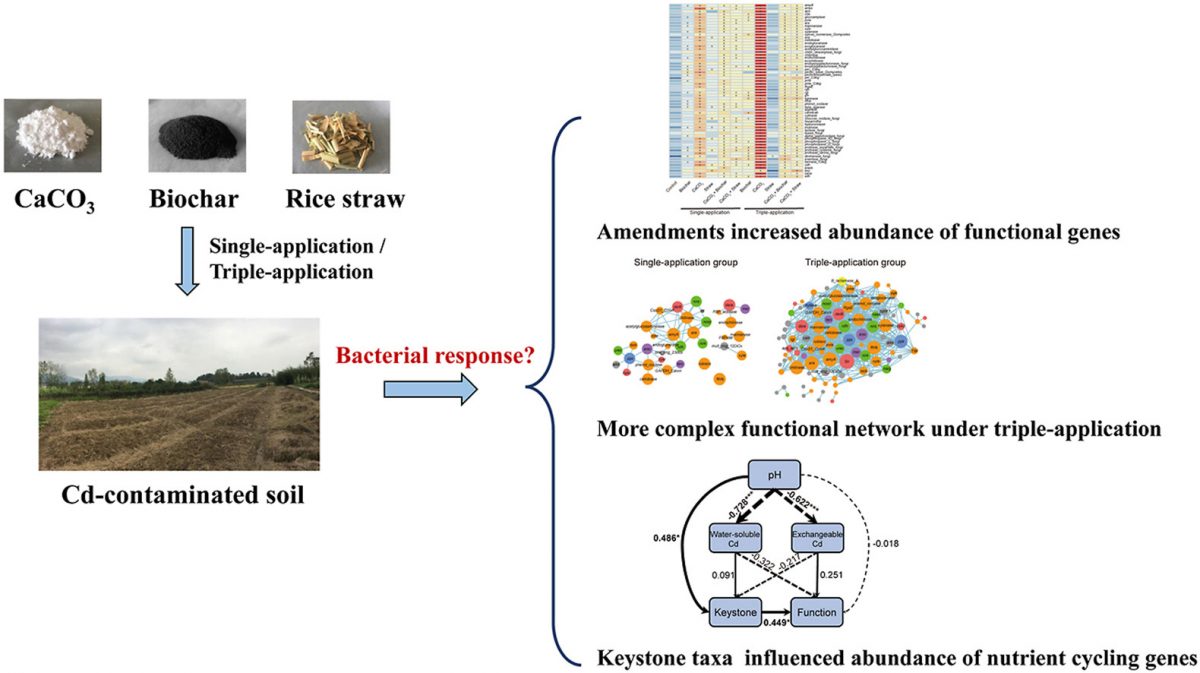
- Repeated application of amendments increased gene abundance of C/N/P cycling.
- Soil pH altered keystone taxa which are highly associated with functional genes.
Abstract
Soil amendments such as lime, biochar and straw are expected to be effective for in situ remediation of cadmium (Cd)-contaminated soil, while increasing the taxonomic diversity of soil microbial community and their functions. The objective of this three-year field experiment was to evaluate the effects of five commonly used soil amendments on Cd stabilization, as well as bacterial phylogenetic and functional attributes. The taxonomic structure of the soil bacterial community was not changed after soil amendments were applied. Most of bacterial functional genes involved in carbon, nitrogen and phosphorus cycling were more abundant in soil amended with CaCO3 and its combination with biochar/straw than soil without amendments. The increase in the number of functional genes was associated with the growth of rare bacterial species, which were 1.3 times more abundant in the CaCO3-amended soil than the unamended soil. Repeated application of CaCO3 and CaCO3 together with biochar/straw increased the number and abundance of functional genes, and the complexity and connectivity of the soil bacterial network, probably because the increased soil pH altered keystone taxa which were highly associated with functional genes. This study demonstrates that soil amendments applied to Cd-contaminated soils can have unexpected consequences on soil bacterial communities and their functions.